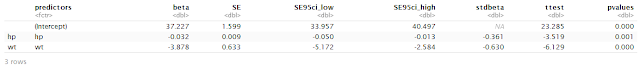
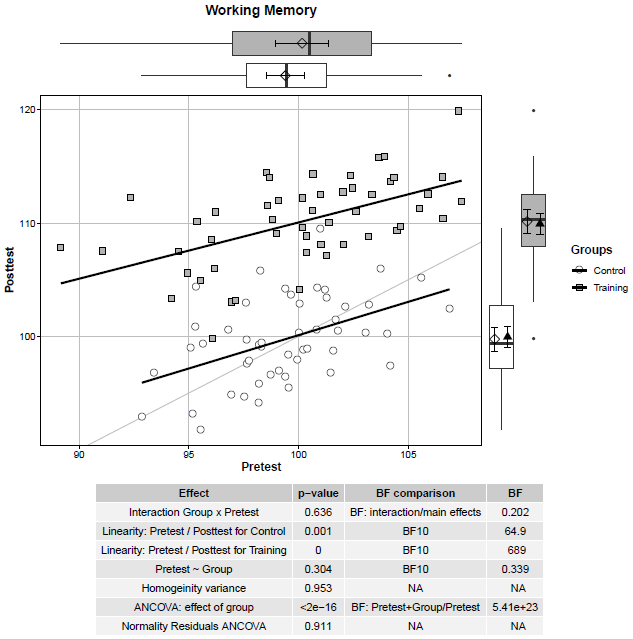
Scientific plots using ggplot2 in R

Below you can find the R code to create a publication-ready plot. You can change the details of the theme() functions to tail the plot to your needs. # Loading required libraries library(ggplot2) library(grid) # Load the function theme_science1 <- function(base_size = 18, facet_border_col = "white", legend_justification = c(0,1), legend_position = c(0,1)){ theme_bw(base_size = base_size) + theme(axis.line = element_line(size = 0.7, color = 'black'), panel.border = element_blank(), panel.grid.major = element_line(size = 0.1, color = "grey90", linetype = "dotted"), ...